
Prof. Ulrich Technau
Vice Dean of Faculty of Life Sciences
Vice Head of Department of Neurosciences and Developmental Biology
Professor for Developmental Biology
BioSketch
Team
Publications
Funding
Vienna cell network

Nematostella vectensis polyp
How did the morphological complexity of Bilateria arise and how is this encoded in the genome?
Which developmental mechanisms and genetic pathways have been preserved throughout evolution and which have been changed to account for changes in morphological complexity?
To address these questions, we mainly focus on the development and genetics of cnidarians. They are the sister group to Bilateria and branched off about 600 Mio years ago. While Bilateria typically have two body axes (anterior-posterior and dorso-ventral axis), a central nervous system and are composed of three germ layers (endoderm, ectoderm and mesoderm), most cnidarians (i.e. the Medusozoans, but not anthozoans) are radially symmetric, they have a diffuse nervous system and the consist of only two cell layers, traditionally referred to as endoderm and ectoderm. Compared to most bilaterians, cnidarians have a stunning regenerative capacity and many are considered virtually immortal, or at least very longlived. Our main model organism is the sea anemone Nematostella vectensis, but we also work on a variety of other cnidarians and non-bilaterians for comparative reasons.
Our current projects aim:
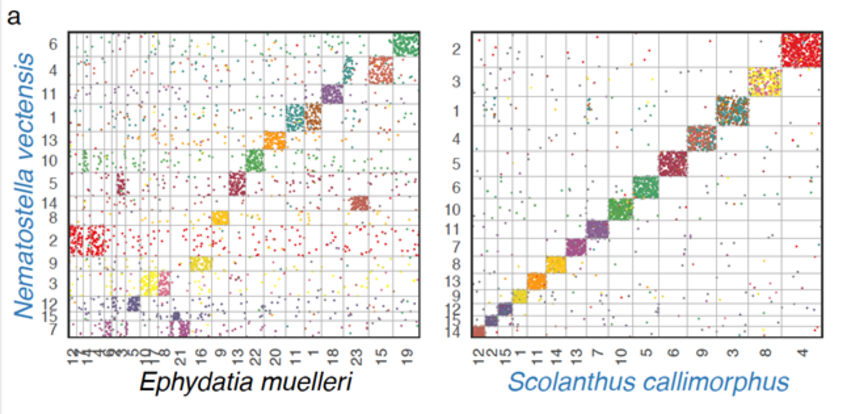
- To understand the genomic architecture and principles of gene regulation. To this end, we have just released a chromosome scale genome assembly and analysed the correlation of presence / absence of topological associated domains (TADs), macro- and microsynteny conservation and location of cis-regulatory elements.
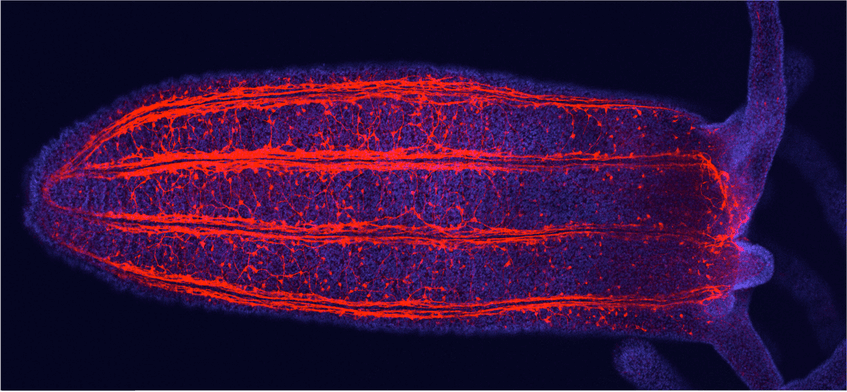
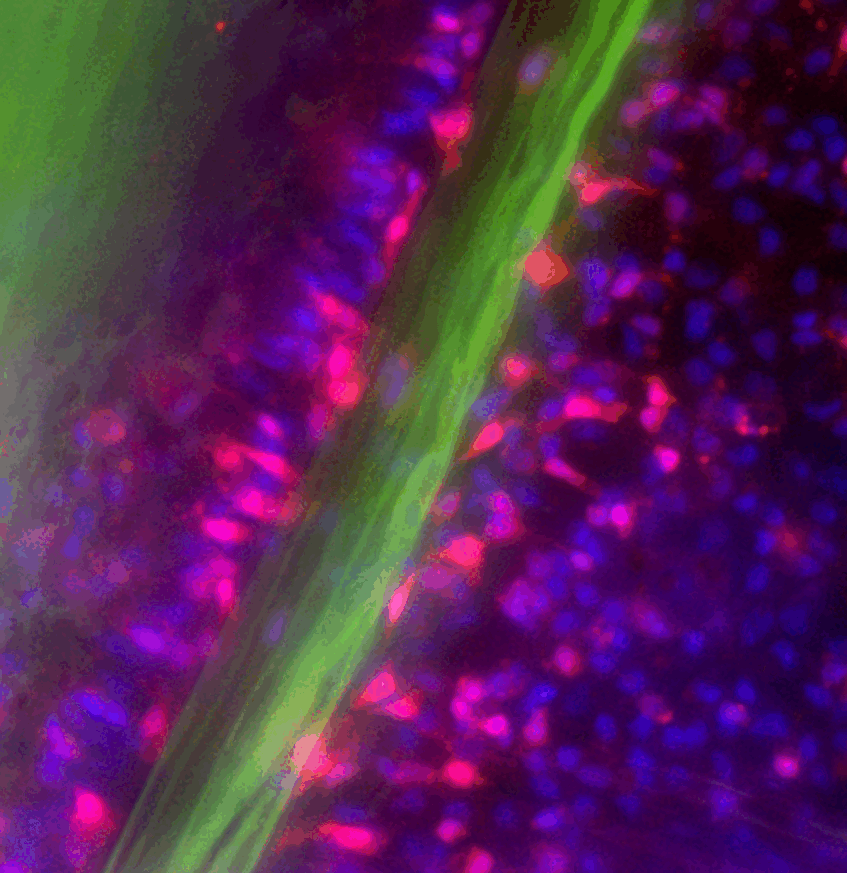
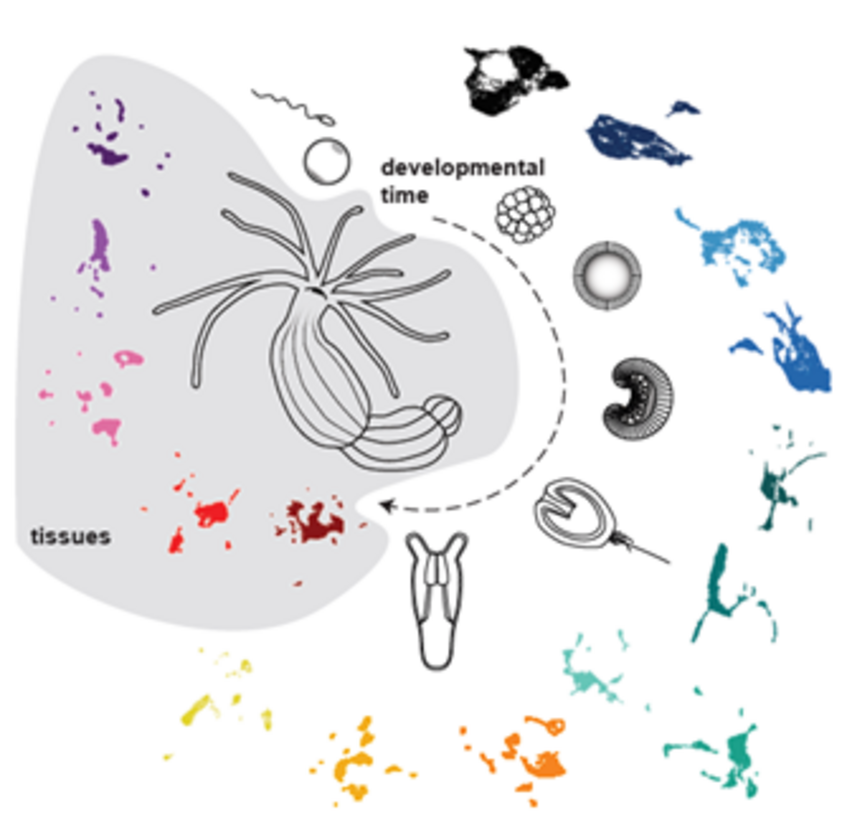
- To understand the structure, complexity and development of the nervous system. To this end, we exploit large single cell RNAseq datasets which helped us to identify many distinct subpopulations of neurons. In a combination of transgenics, CRISPR-mediated mutagenesis and functional genomics, we explore the role of each of them in Nematostella.
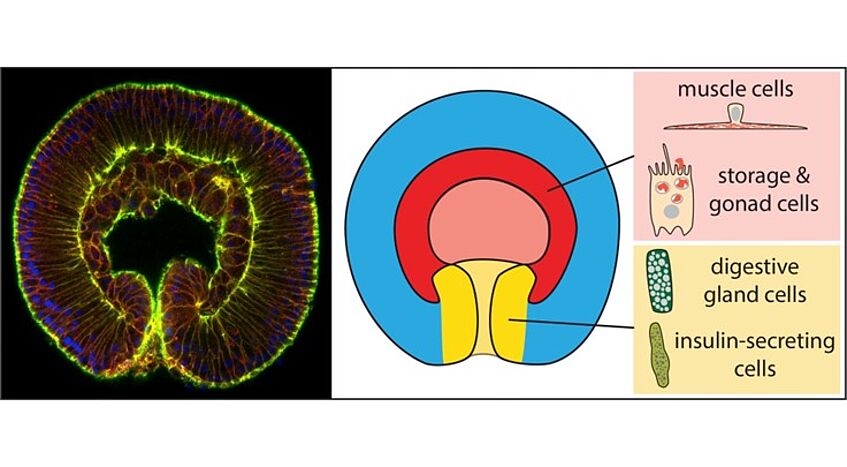
- To understand the evolutionary origin of the three germ layers, we investigate the integration of various signaling pathways demarcating the future germ layer territories and to follow the gene regulatory networks within each of these germ layer territories by multiomics (scRNAseq, scATACseq), ChIP-seq and gene knockdowns /knockouts.
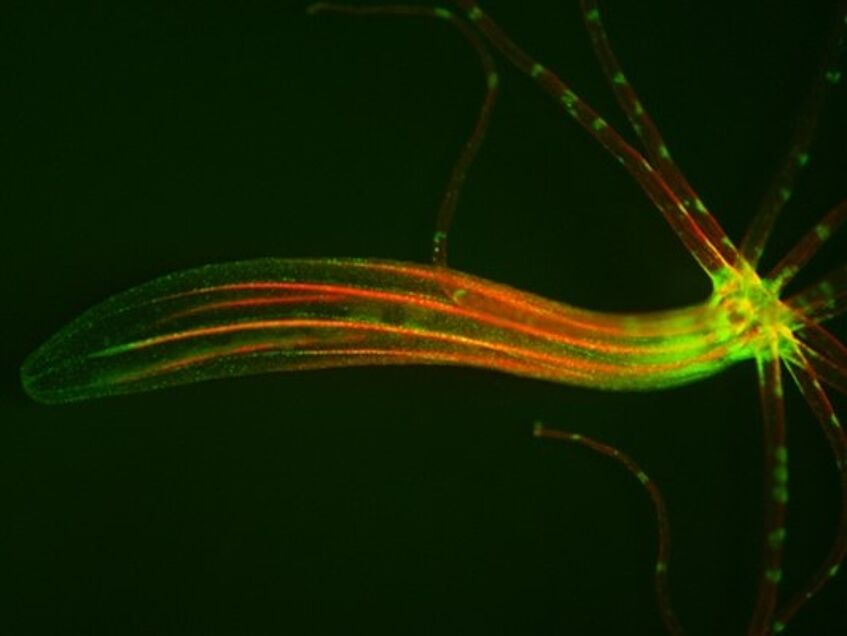
- To understand the evolutionary origin of muscles, the major derivative of mesoderm in Bilateria, we utilize a combination of single cell transcriptomics, transgenics, functional and bioinformatic approaches. This is done in several cnidarian species, including jellyfish which have striated muscles.
- To understand the basis of regeneration and longevity, we aim to identify the multi- and pluripotent stem cells in Nematostella. Candidate cell populations were derived from single cell RNAseq data and validated by transgenics. We have also developed an aggregate system, allowing us to study whole body regeneration by self-organisation. In this system, we can manipulate individual cells and follow their fate.
Selected recent publications
- Zimmermann B, Montenegro JD, Robb SMC, Genikhovich G, Fropf WJ, Weilguny L, He S, Chen S, Lovegrove-Walsh J, Hill EM, Ragkousi K, Praher D, Fredman D, Schultz D, Moran Y, Simakov O, Gibson MC*, Technau U.* (2023).Topological structures and conservation of synteny in sea anemone genomes. Nature Comm, in press. bioRxiv 2020.10.30.359448; doi.org/10.1101/2020.10.30.359448
- Cole AG, Jahnel SM, Kaul S, Steger J, Hagauer, J, Taudes E, Zimmerman B, Reischl R, Denner A, Ferrer Murguia P, Steinmetz P, Technau U. (2023). Muscle cell-type diversification is driven by bHLH transcription factor expansion and facilitated by extensive gene duplications. Nature Comm. 14(1):1747. doi: 10.1038/s41467-023-37220-6.
- Schwaiger M, Andrikou C, Dnyansagar R, Ferrer Murguia P, Paganos P, Voronov D, Zimmermann B, Lebedeva T, Schmidt HA, Genikhovich, G, Benvenuto G, Arnone MI*, Technau U.* (2022). An ancestral Wnt-Brachyury feedback loop in axial patterning and recruitment of mesoderm-determining target genes. Nature Ecol. Evol. 6(12):1921-1939. doi.org/10.1038/s41559-022-01905-w.
- Steger J, Cole AG, Denner A, Lebedeva T, Genikhovich G, Ries A, Reischl R, Taudes E, Lassnig M, Technau U.* (2022). Single cell transcriptomics identifies conserved regulators of neuroglandular lineages. Cell Reports, 40(12):111370. doi: 10.1016/j.celrep.2022.111370.
- Pukhlyakova, E, Aman, A and Technau, U (2019) b-catenin dependent mechanotransduction dates back to the common ancestor of Cnidaria and Bilateria. Proc Natl Acad Sci U S A, 115(24):6231-6236. doi: 10.1073/pnas.1713682115.
- Kirillova, A, Genikhovich, G, Pukhlyakova, E, Demilly, A, Kraus, Y, Technau, U (2018). Germ layer commitment and axis formation in sea anemone embryonic cell aggregates. Proc Natl Acad Sci U S A, 115, 1813-1818.
- Steinmetz, P.R.H.*, Aman, A., Kraus, J.E.M., Technau, U. (2017). Gut-like ectodermal tissue in a sea anemone challenges germ layer homology. Nature Ecol Evol. DOI: 10.1038/s41559-017-0285-5.
